الكود الجيني
الكود الجيني الكود الجيني هو مجموعة من القواعد التى بها تشفر البيانات في داخل المادة الوراثية الدنا أو الرنا (DNA أو RNA sequences) تسلسل وهى ترجمة بيولوجية الى تسلسل البروتينات بواسطة الخلية الحية،والشفرة تحدد خريطة تسلسلية ثلاثية النواة, تسمى "الكودونات" codons والأحماض الأمينية. وهناك تسلسل معلومات النظام الجيني الثلاثي في تسلسل الحمض النووي وعادة ما تحدد واحد من الأحماض الأمينية. على الرغم من ادراج اثنين من الأحماض الأمينية في واحد من الكودونات في تسلسل معلومات النظام الجيني يمكن أن يحدث لبس في أماكن مختلفة في نفس البروتين . [1]. لأن نسبة كبيرة من المورث تشفر بنفس الشفرة تماما (انظر #جدول كودونات الرنا)، هذه الشفرة الخاصة غالبا تعرف بأنها the canonical or standard genetic code, أو ببساطةthe الكود الجيني، وفى الحقيقة توجد اختلافات عن الكود العادي variant codes. Thus the canonical genetic code is not universal. For example, in humans, protein synthesis in mitochondria يعتمد على كود جيني يختلف عن the canonical code. It is important to know that not all المعلومات الجينية is stored as the genetic code. All organisms' DNA contain regulatory sequences, intergenic segments, chromosomal structural areas, which can contribute greatly to phenotype but operate using a distinct sets of rules that may or may not be as straightforward as the codon-to-amino acid paradigm that usually underlies the genetic code.
كسر الشفرة الجينية
بعد أن كسر شيفرة بنية الدنا جيمس واطسون، فرانسيس كريك، موريس ولكنزوروزاليند فرانكلينفإن جهودا جادة لفهم طبيعة تشفير البروتينات قد بدأت.أفترض چورج جامو أن الشفرة من ثلاثة أحرف يجب أن تطبق لتشفير العشرون حامضا أمينيا الأساسيين المستخدمة بواسطة الخلايا الحية لتشفير البروتينات encode the 20 standard amino acids used by living cells to encode proteins is the smallest integer n such that 4n is at least 20هو على الأقل 20 الحقيفة القائلة بأن الكودونات تتكون من 3 قواعد دنا قد كشف عنه Crick, Brenner et al. experiment]]. التوضيح الأولى ل codon قد تم بواسطة Marshall Nirenberg و Heinrich J. Matthaei في عام 1961 في National Institutes of Health .وقد أستخدمواcell-free system لتشفير poly-uracil RNA sequence (أو UUUUU... من وجهة بيو كيميائية) و أكتشفوا أن البولي بيبتايد الذي قامو بتخليقه, يحتوى فقط على الحمض الأميني phenylalanine. وهم بالتالى أستنتجوا أن البروتين المتعدد الفينيل ألانين أن الكودون UUU قد خصص الحمض الأمينى فينيل ألانين. وبتوسيع هذا الجهد فإن Nirenberg وفريقه كانوا قادرين على تعيين nucleotide makeup لكل codon. ومن أجل تعيين الترتيب التسلسلي فإن trinucleotides were bound to ribosomeو radioactively labeled aminoacyl-tRNA was used to determine which amino acid corresponded to the codon. Nirenberg's group was able to determine the sequences of 54 out of 64 codons.
Subsequent work by هار گوبـِند خورانا identified the rest of the code, and shortly thereafter Robert W. Holley determined the structure of transfer RNA, the adapter molecule that facilitates translation. This work was based upon earlier studies by Severo Ochoa, who received the Nobel prize in 1959 for his work on the enzymology of RNA synthesis. In 1968, Khorana, Holley and Nirenberg also received جائزة نوبل في الفسيولوجيا أو الطب لعملهم.
انتقال المعلومات عن طريق الكود الجيني
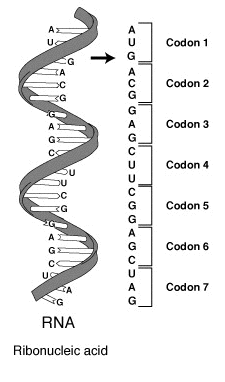
جينوم أي عضية is inscribed in دنا, or in some viruses RNA. The portion of the genome that codes for a protein or an RNA is referred to as a مورثة. Those genes that code for proteins are composed of tri-nucleotide units called codons, each coding for a single amino acid. Each nucleotide sub-unit consists of a فوسفات, deoxyribose sugar and one of the 4 nitrogenous nucleotide bases. The پيورين bases أدينين (A) وجوانين (G) هم أكبر ويتكونون من 2 حلقة عطرية The pyrimidine bases cytosine (C) and thymine (T) هم أصغرويتكونون من حلقة عطرية واحدة فقط . In the double-helix configuration, two strands of DNA ويرتبطون ببعضهم بواسطة رابطة هيدروجين بترتيب يطلق عليه base pairing. هذه الروابط دائما تكون بين adenine base on one strand and a thymine على الآخر strand و بين a cytosine base on one strand and a guanine base on the other. This means that the number of A and T residues will be the same in a given double helix, as will the number of G and C residues. In RNA, thymine (T) is replaced by uracil (U), and the deoxyribose is substituted by ribose. كل جين(كود أو شفرة)مرمزة البروتين هو transcribed الى قالب جزيئ لل polymer RNA المقابل يعرف بالرنا الرسول messenger RNA ormRNA]] هو بدوره translated على ال ribosome into an amino acid chain or polypeptide. إن عملية الترجمة تتطلب transfer RNAs المحدد لكل حمض نووى مع كل حمض نووى ملتصق بهمguanosine triphosphate كمصدر للطاقة,and a number of transla factors tRNAs have anticodons complementary to the codons in mRNA and can be charged covalently with [[amino acids] at their 3' terminal CCA ends. Individual tRNAs are charge with specific amino acids by enzymes known as aminoacyl tRNA synthetases, which have high specificity for both their cognate amino acids and tRNAs. The high specificity of these enzymes is a major reason why the fidelity of protein translation is maintained
are 4³ = 64 different codon combinations possible with a triplet codon of three nucleotides. In reality, all 64 codons of the standard genetic code are assigned for either amino acids or stop signals during translation. If, for example, an RNA sequence, UUUAAACCC is considered and the reading-frame starts with the first U (by convention, 5' to 3'), there are three codons, namely, UUU, AAA and CCC, each of which specifies one amino acid. This RNA sequence will be translated into an amino acid sequence, three amino acids long. A comparison may be made with computer science, where the codon is the equivalent of a word, which is the standard "chunk" for handling data (like one amino acid of a protein), and a nucleotide for a bit.
The standard genetic code is shown in the following tables. Table 1 shows what amino acid each of the 64 codons specifies. Table 2 shows what codons specify each of the 20 standard amino acids involved in translation. These are called forward and reverse codon tables, respectively. For example, the codon AAU represents the amino acid asparagine, and UGU and UGC represent cysteine (standard three-letter designations, Asn and Cys, respectively).
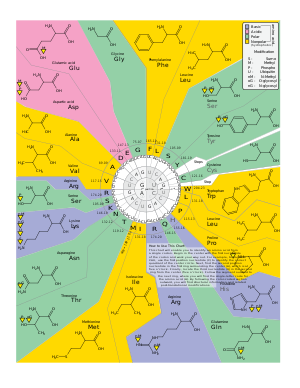
جدول كودونات الرنا
| غير قطبي | قطبي | قاعدي | حامضي |
| القاعدة الثانية | |||||
|---|---|---|---|---|---|
| U | C | A | G | ||
| القاعدة الأولى |
U | UUU (Phe/F) Phenylalanine UUC (Phe/F) Phenylalanine |
UCU (Ser/S) Serine UCC (Ser/S) Serine |
UAU (Tyr/Y) Tyrosine UAC (Tyr/Y) Tyrosine |
UGU (Cys/C) Cysteine UGC (Cys/C) Cysteine |
| UUA (Leu/L) Leucine | UCA (Ser/S) Serine | UAA Ochre (Stop) | UGA Opal (Stop) | ||
| UUG (Leu/L) Leucine | UCG (Ser/S) Serine | UAG Amber (Stop) | UGG (Trp/W) Tryptophan | ||
| C | CUU (Leu/L) Leucine CUC (Leu/L) Leucine |
CCU (Pro/P) Proline CCC (Pro/P) Proline |
CAU (His/H) Histidine CAC (His/H) Histidine |
CGU (Arg/R) Arginine CGC (Arg/R) Arginine | |
| CUA (Leu/L) Leucine CUG (Leu/L) Leucine |
CCA (Pro/P) Proline CCG (Pro/P) Proline |
CAA (Gln/Q) Glutamine
CAG (Gln/Q) Glutamine |
CGA (Arg/R) Arginine CGG (Arg/R) Arginine | ||
| A | AUU (Ile/I) Isoleucine AUC (Ile/I) Isoleucine |
ACU (Thr/T) Threonine ACC (Thr/T) Threonine |
AAU (Asn/N) Asparagine AAC (Asn/N) Asparagine |
AGU (Ser/S) Serine AGC (Ser/S) Serine | |
| AUA (Ile/I) Isoleucine | ACA (Thr/T) Threonine | AAA (Lys/K) Lysine | AGA (Arg/R) Arginine | ||
| AUG (Met/M) Methionine, Start[2] |
ACG (Thr/T) Threonine | AAG (Lys/K) Lysine | AGG (Arg/R) Arginine | ||
| G | GUU (Val/V) Valine GUC (Val/V) Valine |
GCU (Ala/A) Alanine GCC (Ala/A) Alanine |
GAU (Asp/D) Aspartic acid GAC (Asp/D) Aspartic acid |
GGU (Gly/G) Glycine GGC (Gly/G) Glycine | |
| GUA (Val/V) Valine GUG (Val/V) Valine |
GCA (Ala/A) Alanine GCG (Ala/A) Alanine |
GAA (Glu/E) Glutamic acid GAG (Glu/E) Glutamic acid |
GGA (Gly/G) Glycine GGG (Gly/G) Glycine | ||
| Ala/A | GCU, GCC, GCA, GCG | Leu/L | UUA, UUG, CUU, CUC, CUA, CUG |
|---|---|---|---|
| Arg/R | CGU, CGC, CGA, CGG, AGA, AGG | Lys/K | AAA, AAG |
| Asn/N | AAU, AAC | Met/M | AUG |
| Asp/D | GAU, GAC | Phe/F | UUU, UUC |
| Cys/C | UGU, UGC | Pro/P | CCU, CCC, CCA, CCG |
| Gln/Q | CAA, CAG | Ser/S | UCU, UCC, UCA, UCG, AGU, AGC |
| Glu/E | GAA, GAG | Thr/T | ACU, ACC, ACA, ACG |
| Gly/G | GGU, GGC, GGA, GGG | Trp/W | UGG |
| His/H | CAU, CAC | Tyr/Y | UAU, UAC |
| Ile/I | AUU, AUC, AUA | Val/V | GUU, GUC, GUA, GUG |
| START | AUG | STOP | UAG, UGA, UAA |
Salient features
Reading frame of a sequence
Note that a codon is defined by the initial nucleotide from which translation starts. For example, the string GGGAAACCC, if read from the first position, contains the codons GGG, AAA and CCC; and, if read from the second position, it contains the codons GGA and AAC; if read starting from the third position, GAA and ACC. Partial codons have been ignored in this example. Every sequence can thus be read in three reading frames, each of which will produce a different amino acid sequence (in the given example, Gly-Lys-Pro, Gly-Asp, or Glu-Thr, respectively). With double-stranded DNA there are six possible reading frames, three in the forward orientation on one strand and three reverse (on the opposite strand).
The actual frame in which a protein sequence is translated is defined by a start codon, usually the first AUG codon in the mRNA sequence. Mutations that disrupt the reading frame by insertions or deletions of a non-multiple of 3 nucleotide bases are known as frameshift mutations. These mutations may impair the function of the resulting protein, if it is formed, and are thus rare in in vivo protein-coding sequences. Often such misformed proteins are targeted for proteolytic degradation. In addition, a frame shift mutation is very likely to cause a stop codon to be read, which truncates the creation of the protein (example [2]). One reason for the rareness of frame-shifted mutations' being inherited is that, if the protein being translated is essential for growth under the selective pressures the organism faces, absence of a functional protein may cause lethality before the organism is viable.
كودونات البداية/التوقف
Translation starts with a chain initiation codon (start codon). Unlike stop codons, the codon alone is not sufficient to begin the process. Nearby sequences (Such as the Shine-Dalgarno sequence in E.Coli) and initiation factors are also required to start translation. The most common start codon is AUG, which also codes for methionine. There are sometimes other alternative start codons (depending on the organism), such as "UUG," which normally codes for leucine. However, when used as a start codon, these alternative start codons are usually translated as methionine (regardless of their normal meaning).[3]
The three stop codons have been given names: UAG is amber, UGA is opal (sometimes also called umber), and UAA is ochre. "Amber" was named by discoverers Richard Epstein and Charles Steinberg after their friend Harris Bernstein, whose last name means "amber" in German. The other two stop codons were named 'ochre" and "opal" in order to keep the "color names" theme. Stop codons are also called termination codons and they signal release of the nascent polypeptide from the ribosome due to binding of release factors in the absence of cognate tRNAs with anticodons complementary to these stop signals.[4]
Degeneracy of the genetic code
The genetic code has redundancy but no ambiguity (see the codon tables above for the full correlation). For example, although codons GAA and GAG both specify glutamic acid (redundancy), neither of them specifies any other amino acid (no ambiguity). The codons encoding one amino acid may differ in any of their three positions. For example the amino acid glutamic acid is specified by GAA and GAG codons (difference in the third position), the amino acid leucine is specified by UUA, UUG, CUU, CUC, CUA, CUG codons (difference in the first or third position), while the amino acid serine is specified by UCA, UCG, UCC, UCU, AGU, AGC (difference in the first, second or third position).
A position of a codon is said to be a fourfold degenerate site if any nucleotide at this position specifies the same amino acid. For example, the third position of the glycine codons (GGA, GGG, GGC, GGU) is a fourfold degenerate site, because all nucleotide substitutions at this site are synonymous; i.e., they do not change the amino acid. Only the third positions of some codons may be fourfold degenerate. A position of a codon is said to be a twofold degenerate site if only two of four possible nucleotides at this position specify the same amino acid. For example, the third position of the glutamic acid codons (GAA, GAG) is a twofold degenerate site. In twofold degenerate sites, the equivalent nucleotides are always either two purines (A/G) or two pyrimidines (C/U), so only transversional substitutions (purine to pyrimidine or pyrimidine to purine) in twofold degenerate sites are nonsynonymous. A position of a codon is said to be a non-degenerate site if any mutation at this position results in amino acid substitution. There is only one threefold degenerate site where changing three of the four nucleotides has no effect on the amino acid, while changing the fourth possible nucleotide results in an amino acid substitution. This is the third position of an isoleucine codon: AUU, AUC, or AUA all encode isoleucine, but AUG encodes methionine. In computation this position is often treated as a twofold degenerate site.
There are three amino acids encoded by six different codons: serine, leucine, arginine. Only two amino acids are specified by a single codon; one of these is the amino-acid methionine, specified by the codon AUG, which also specifies the start of translation; the other is tryptophan, specified by the codon UGG. The degeneracy of the genetic code is what accounts for the existence of silent mutations.
Degeneracy results because a triplet code designates 20 amino acids and a stop codon. Because there are four bases, triplet codons are required to produce at least 21 different codes. For example, if there were two bases per codon, then only 16 amino acids could be coded for (4²=16). Because at least 21 codes are required, then 4³ gives 64 possible codons, meaning that some degeneracy must exist.
These properties of the genetic code make it more fault-tolerant for point mutations. For example, in theory, fourfold degenerate codons can tolerate any point mutation at the third position, although codon usage bias restricts this in practice in many organisms; twofold degenerate codons can tolerate one out of the three possible point mutations at the third position. Since transition mutations (purine to purine or pyrimidine to pyrimidine mutations) are more likely than transversion (purine to pyrimidine or vice-versa) mutations, the equivalence of purines or that of pyrimidines at twofold degenerate sites adds a further fault-tolerance.
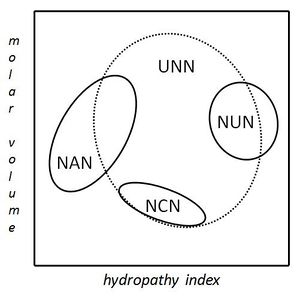
A practical consequence of redundancy is that some errors in the genetic code only cause a silent mutation or an error that would not affect the protein because the hydrophilicity or hydrophobicity is maintained by equivalent substitution of amino acids; for example, a codon of NUN (where N = any nucleotide) tends to code for hydrophobic amino acids. NCN yields amino acid residues that are small in size and moderate in hydropathy; NAN encodes average size hydrophilic residues; UNN encodes residues that are not hydrophilic.[5][6] These tendencies may result from that the aminoacyl tRNA synthetases related the such codons share a common ancestry.
Even so, single point mutations can still cause dysfunctional proteins. For example, a mutated hemoglobin gene causes sickle-cell disease. In the mutant hemoglobin a hydrophilic glutamate (Glu) is substituted by the hydrophobic valine (Val), which reduces the solubility of β-globin. In this case, this mutation causes hemoglobin to form linear polymers linked by the hydrophobic interaction between the valine groups causing sickle-cell deformation of erythrocytes. Sickle-cell disease is generally not caused by a de novo mutation. Rather it is selected for in malarial regions (in a way similar to thalassemia), as heterozygous people have some resistance to the malarial Plasmodium parasite (heterozygote advantage).
These variable codes for amino acids are allowed because of modified bases in the first base of the anticodon of the tRNA, and the base-pair formed is called a wobble base pair. The modified bases include inosine and the Non-Watson-Crick U-G basepair.
تنويعات على الكود الجيني القياسي
While slight variations on the standard code had been predicted earlier,[7] none were discovered until 1979, when researchers studying human mitochondrial genes discovered they used an alternative code. Many slight variants have been discovered since,[8] including various alternative mitochondrial codes,[9] as well as small variants such as Mycoplasma translating the codon UGA as tryptophan. In bacteria and archaea, GUG and UUG are common start codons. However, in rare cases, certain specific proteins may use alternative initiation (start) codons not normally used by that species.[10]
In certain proteins, non-standard amino acids are substituted for standard stop codons, depending upon associated signal sequences in the messenger RNA: UGA can code for selenocysteine and UAG can code for pyrrolysine as discussed in the relevant articles. Selenocysteine is now viewed as the 21st amino acid, and pyrrolysine is viewed as the 22nd. A detailed description of variations in the genetic code can be found at the NCBI web site.
Notwithstanding these differences, all known codes have strong similarities to each other, and the coding mechanism is the same for all organisms: three-base codons, tRNA, ribosomes, reading the code in the same direction and translating the code three letters at a time into sequences of amino acids.
نظريات عن أصل الكود الجيني
Despite the variations that exist, the genetic codes used by all known forms of life on Earth are very similar. Since there are many possible genetic codes that are thought to have similar utility to the one used by Earth life, the theory of evolution suggests that the genetic code was established very early in the history of life. Phylogenetic analysis of transfer RNA suggests that tRNA molecules evolved before the present set of aminoacyl-tRNA synthetases.[11]
The genetic code is not a random assignment of codons to amino acids.[12] For example, amino acids that share the same biosynthetic pathway tend to have the same first base in their codons,[13] and amino acids with similar physical properties tend to have similar codons.[14][15]
There are three themes running through the many theories that seek to explain the evolution of the genetic code (and hence the origin of these patterns).[16]:
- Recent aptamer experiments show that some amino acids have a selective chemical affinity for the base triplets that code for them.[17] This suggests that the current complex translation mechanism involving tRNA and associated enzymes may be a later development, and that originally, protein sequences were directly templated on base sequences.
- That the standard modern genetic code grew from a simpler earlier code through a process of "biosynthetic expansion". Here the idea is that primordial life 'discovered' new amino acids (e.g., as by-products of metabolism) and later back-incorporated some of these into the machinery of genetic coding. Although much circumstantial evidence has been found to suggest that fewer different amino acids were used in the past than today,[18] precise and detailed hypotheses about exactly which amino acids entered the code in exactly what order has proved far more controversial.[19][20]
- That natural selection has led to codon assignments of the genetic code that minimize the effects of mutations.[21]
المصادر
- ^ http://www.sciencemag.org/cgi/content/short/323/5911/259
- ^ The codon AUG both codes for methionine and serves as an initiation site: the first AUG in an mRNA's coding region is where translation into protein begins.
- ^ Touriol C, Bornes S, Bonnal S; et al. (2003). "Generation of protein isoform diversity by alternative initiation of translation at non-AUG codons". Biology of the cell / under the auspices of the European Cell Biology Organization. 95 (3–4): 169–78. PMID 12867081.
{{cite journal}}: Explicit use of et al. in:|author=(help)CS1 maint: multiple names: authors list (link) - ^ How nonsense mutations got their names
- ^ Yang et al. 1990. In Reaction Centers of Photosynthetic Bacteria. M.-E. Michel-Beyerle. (Ed.) (Springer-Verlag, Germany) 209-218
- ^ Genetic Algorithms and Recursive Ensemble Mutagenesis in Protein Engineering http://www.complexity.org.au/ci/vol01/fullen01/html/
- ^ Crick, F. H. C. and Orgel, L. E. (1973) "Directed panspermia." Icarus 19:341-346. p. 344: "It is a little surprising that organisms with somewhat different codes do not coexist." (Further discussion at [1])
- ^ NCBI: "The Genetic Codes", Compiled by Andrzej (Anjay) Elzanowski and Jim Ostell
- ^ Jukes TH, Osawa S, The genetic code in mitochondria and chloroplasts., Experientia. 1990 Dec 1;46(11-12):1117-26.
- ^ Genetic Code page in the NCBI Taxonomy section (Downloaded 27 April 2007.)
- ^ De Pouplana, L.R. (1998). "Genetic code origins: tRNAs older than their synthetases?". Proceedings of the National Academy of Sciences. 95 (19): 11295. doi:10.1073/pnas.95.19.11295. PMID 9736730.
{{cite journal}}: Unknown parameter|coauthors=ignored (|author=suggested) (help) - ^ Freeland SJ, Hurst LD (1998). "The genetic code is one in a million". J. Mol. Evol. 47 (3): 238–48. doi:10.1007/PL00006381. PMID 9732450.
{{cite journal}}: Unknown parameter|month=ignored (help) - ^ Taylor FJ, Coates D (1989). "The code within the codons". BioSystems. 22 (3): 177–87. doi:10.1016/0303-2647(89)90059-2. PMID 2650752.
- ^ Di Giulio M (1989). "The extension reached by the minimization of the polarity distances during the evolution of the genetic code". J. Mol. Evol. 29 (4): 288–93. doi:10.1007/BF02103616. PMID 2514270.
{{cite journal}}: Unknown parameter|month=ignored (help) - ^ Wong JT (1980). "Role of minimization of chemical distances between amino acids in the evolution of the genetic code". Proc. Natl. Acad. Sci. U.S.A. 77 (2): 1083–6. doi:10.1073/pnas.77.2.1083. PMID 6928661.
{{cite journal}}: Unknown parameter|month=ignored (help) - ^ Knight, R.D.; Freeland S. J. and Landweber, L.F. (1999) The 3 Faces of the Genetic Code. Trends in the Biochemical Sciences 24(6), 241-247.
- ^ Knight, R.D. and Landweber, L.F. (1998). Rhyme or reason: RNA-arginine interactions and the genetic code. Chemistry & Biology 5(9), R215-R220. PDF version of manuscript
- ^ Brooks, Dawn J.; Fresco, Jacques R.; Lesk, Arthur M.; and Singh, Mona. (2002). Evolution of Amino Acid Frequencies in Proteins Over Deep Time: Inferred Order of Introduction of Amino Acids into the Genetic Code. Molecular Biology and Evolution 19, 1645-1655.
- ^ Amirnovin R. (1997) An analysis of the metabolic theory of the origin of the genetic code. Journal of Molecular Evolution 44(5), 473-6.
- ^ Ronneberg T.A.; Landweber L.F. and Freeland S.J. (2000) Testing a biosynthetic theory of the genetic code: Fact or artifact? Proceedings of the National Academy of Sciences, USA 97(25), 13690-13695.
- ^ Freeland S.J.; Wu T. and Keulmann N. (2003) The Case for an Error Minimizing Genetic Code. Orig Life Evol Biosph. 33(4-5), 457-77.
قراءات اضافية
- Griffiths, Anthony J.F.; Miller, Jeffrey H.; Suzuki, David T.; Lewontin, Richard C.; Gelbart, William M. (1999). Introduction to Genetic Analysis (7th ed.). New York: W. H. Freeman & Co. ISBN 0-7167-3771-X
- Alberts, Bruce; Johnson, Alexander; Lewis, Julian; Raff, Martin; Roberts, Keith; Walter, Peter. (2002). Molecular Biology of the Cell (4th ed.). New York: Garland Publishing. ISBN 0-8153-3218-1
- Lodish, Harvey; Berk, Arnold; Zipursky, S. Lawrence; Matsudaira, Paul; Baltimore, David; Darnell, James E. (1999). Molecular Cell Biology (4th ed.). New York: W. H. Freeman & Co. ISBN 0-7167-3706-X
وصلات خارجية
- The Genetic Codes → Genetic Code Tables
- Online DNA → Amino Acid Converter
- Online DNA Sequence → Protein Sequence converter
- Online DNA to protein translation (6 frames/17+ genetic codes)
- The Codon Usage Database → Codon frequency tables for many organisms
- Amino Acid Tree - DNA Code to Amino Acid Reference