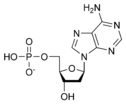
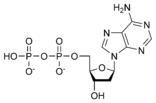
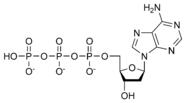
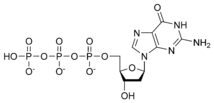
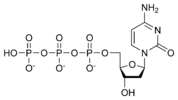
نيوكليوتيد
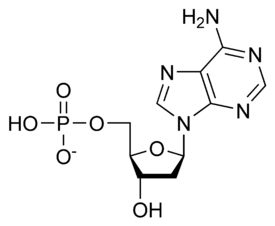
نيوكليوتايد Nucleotide هي الوحدة اساسية لبناء الدنا و الرنا. في الأحماض النووية الوراثية (دنا و رنا) تتكون وحدة النيوكليوتيد من:
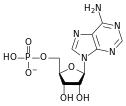
- سكر خماسي (Ribose Sugar) وفي حمض الدنا يكون هذا السكر منقوص الأكسجين.
- مجموعة فوسفات.
- قاعدة نيتروجينية، في حمض الدنا القواعد النيتروجينية هي أدينين (Adenine) و كوانين (Guanine) و ثايمين (Thymine) و سايتوسين (Cytosine). أم في حمض الرنا فيكون اليوراسيل (Uracil) بدل من الثايمين.
البنية
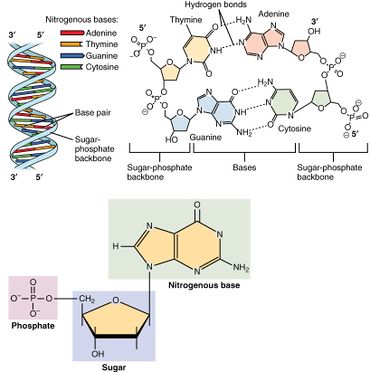
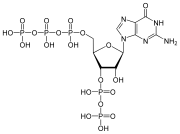
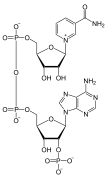
A nucleotide is composed of three distinctive chemical sub-units: a five-carbon sugar molecule, a nitrogenous base—which two together are called a nucleoside—and one phosphate group. With all three joined, a nucleotide is also termed a "nucleoside monophosphate", "nucleoside diphosphate" or "nucleoside triphosphate", depending on how many phosphates make up the phosphate group.
In nucleic acids, nucleotides contain either a purine or a pyrimidine base—i.e., the nitrogenous base molecule, also known as a nucleobase—and are termed ribonucleotides if the sugar is ribose, or deoxyribonucleotides if the sugar is deoxyribose. Individual phosphate molecules repetitively connect the sugar-ring molecules in two adjacent nucleotide monomers, thereby connecting the nucleotide monomers of a nucleic acid end-to-end into a long chain. These chain-joins of sugar and phosphate molecules create a 'backbone' strand for a single- or double helix. In any one strand, the chemical orientation (directionality) of the chain-joins runs from the 5'-end to the 3'-end (read: 5 prime-end to 3 prime-end)—referring to the five carbon sites on sugar molecules in adjacent nucleotides. In a double helix, the two strands are oriented in opposite directions, which permits base pairing and complementarity between the base-pairs, all which is essential for replicating or transcribing the encoded information found in DNA.
Nucleic acids then are polymeric macromolecules assembled from nucleotides, the monomer-units of nucleic acids. The purine bases adenine and guanine and pyrimidine base cytosine occur in both DNA and RNA, while the pyrimidine bases thymine (in DNA) and uracil (in RNA) occur in just one. Adenine forms a base pair with thymine with two hydrogen bonds, while guanine pairs with cytosine with three hydrogen bonds.
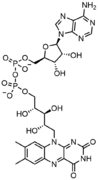
In addition to being building blocks for construction of nucleic acid polymers, singular nucleotides play roles in cellular energy storage and provision, cellular signaling, as a source of phosphate groups used to modulate the activity of proteins and other signaling molecules, and as enzymatic cofactors, often carrying out redox reactions. Signaling cyclic nucleotides are formed by binding the phosphate group twice to the same sugar molecule, bridging the 5'- and 3'- hydroxyl groups of the sugar.[1] Some signaling nucleotides differ from the standard single-phosphate group configuration, in having multiple phosphate groups attached to different positions on the sugar.[2] Nucleotide cofactors include a wider range of chemical groups attached to the sugar via the glycosidic bond, including nicotinamide and flavin, and in the latter case, the ribose sugar is linear rather than forming the ring seen in other nucleotides.
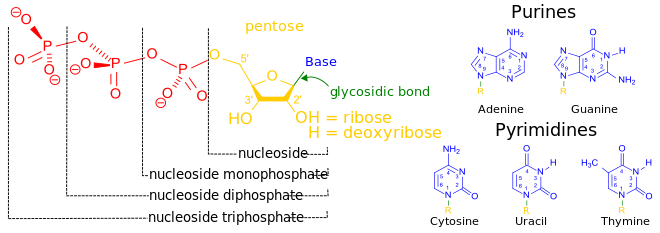
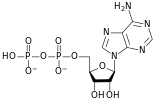
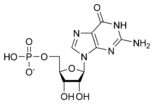
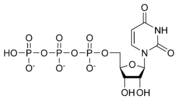
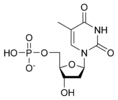
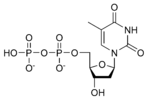
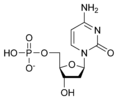
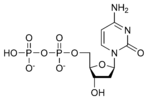
المخططات البنيوية للنيوكليوتيد
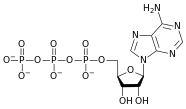
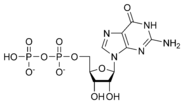
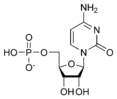
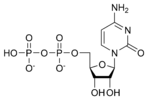
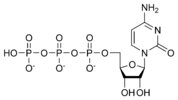
المخططات البنيوية للـ دىأوكسينيوكليوتيد
التخليق
Nucleotides can be synthesized by a variety of means both in vitro and in vivo.
In vitro, protecting groups may be used during laboratory production of nucleotides. A purified nucleoside is protected to create a phosphoramidite, which can then be used to obtain analogues not found in nature and/or to synthesize an oligonucleotide.
In vivo, nucleotides can be synthesized de novo or recycled through salvage pathways.[3] The components used in de novo nucleotide synthesis are derived from biosynthetic precursors of carbohydrate and amino acid metabolism, and from ammonia and carbon dioxide. The liver is the major organ of de novo synthesis of all four nucleotides. De novo synthesis of pyrimidines and purines follows two different pathways. Pyrimidines are synthesized first from aspartate and carbamoyl-phosphate in the cytoplasm to the common precursor ring structure orotic acid, onto which a phosphorylated ribosyl unit is covalently linked. Purines, however, are first synthesized from the sugar template onto which the ring synthesis occurs. For reference, the syntheses of the purine and pyrimidine nucleotides are carried out by several enzymes in the cytoplasm of the cell, not within a specific organelle. Nucleotides undergo breakdown such that useful parts can be reused in synthesis reactions to create new nucleotides.
Pyrimidine ribonucleotide synthesis
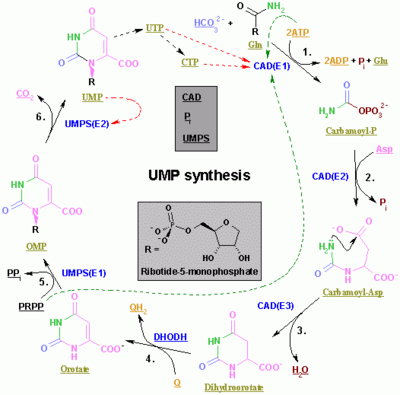
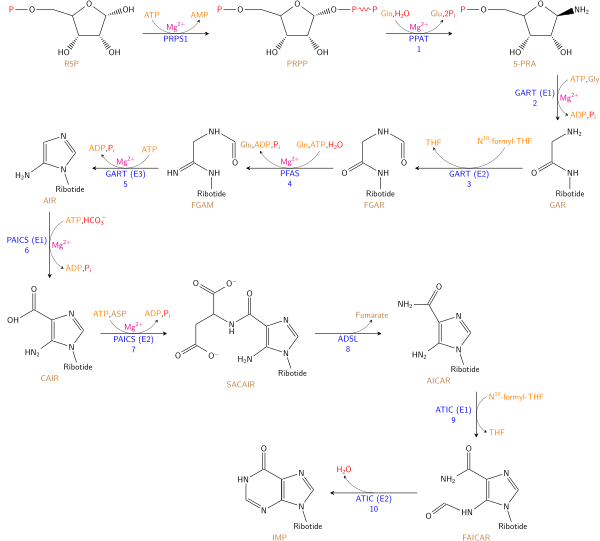
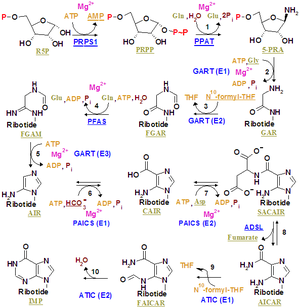
Purine ribonucleotide synthesis
The atoms that are used to build the purine nucleotides come from a variety of sources:
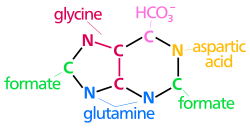 |
The biosynthetic origins of purine ring atoms N1 arises from the amine group of Asp C2 and C8 originate from formate N3 and N9 are contributed by the amide group of Gln C4, C5 and N7 are derived from Gly C6 comes from HCO3− (CO2) |
أنواع القواعد
Nucleotides can be synthesized with both purine and pyrimidine as bases. In DNA, the purine bases are adenine and guanine, while the pyrimidines are thymine and cytosine. RNA uses uracil rather than thymine (thymine is produced by adding a methyl to uracil). The nucleotide passes through numerous biochemical steps while being processed, adding and removing atoms through the use of numerous enzymes.
Pyrimidine ribonucleotides
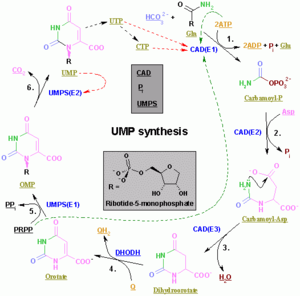
The synthesis of a single pyrimidine is complex; the diagram to the left demonstrates the synthesis of a single pyrimidine.
Purine ribonucleotides
The atoms which are used to build the purine nucleotides come from a variety of sources:
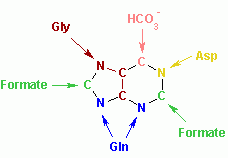 |
The biosynthetic origins of purine ring atoms N1 arises from the amine group of Asp C2 and C8 originate from formate N3 and N9 are contributed by the amide group of Gln C4, C5 and N7 are derived from Gly C6 comes from HCO3- (CO2) |
The de novo synthesis of purine nucleotides by which these precursors are incorporated into the purine ring, proceeds by a 10 step pathway to the branch point intermediate IMP, the nucleotide of the base hypoxanthine. AMP and GMP are subsequently synthesized from this intermediate via separate, two step each, pathways. Thus purine moieties are initially formed as part of the ribonucleotides rather than as free bases.
Six enzymes take part in IMP synthesis. Three of them are multifunctional:
Reaction 1. The pathway starts with the formation of PRPP. PRPS1 is the enzyme that activates R5P, which is primarily formed by the pentose phosphate pathway, to PRPP by reacting it with ATP. The reaction is unusual in that a pyrophosphoryl group is directly transferred from ATP to C1 of R5P and that the product has the α configuration about C1. This reaction is also shared with the pathways for the synthesis of the pyrimidine nucleotides, Trp, and His. As a result of being on (a) such (a) major metabolic crossroad and the use of energy, this reaction is highly regulated.
Reaction 2. In the first reaction unique to purine nucleotide biosynthesis, PPAT catalyzes the displacement of PRPP's pyrophosphate group (PPi) by Gln's amide nitrogen. The reaction occurs with the inversion of configuration about ribose C1, thereby forming β-5-phosphorybosylamine (5-PRA) and establishing the anomeric form of the future nucleotide. This reaction which is driven to completion by the subsequent hydrolysis of the released PPi, is the pathway's flux generating step and is therefore regulated too.
انظر أيضاً
وصلات خارجية
- Abbreviations and Symbols for Nucleic Acids, Polynucleotides and their Constituents (IUPAC)
- Provisional Recommendations 2004 (IUPAC)
- Chemistry explanation of nucleotide structure
- ^ خطأ استشهاد: وسم
<ref>غير صحيح؛ لا نص تم توفيره للمراجع المسماةAlberts - ^ Smith, A. D., ed. (2000). Oxford Dictionary of Biochemistry and Molecular Biology, Revised edition. Oxford: Oxford University Press. p. 460.
- ^ Zaharevitz DW, Anderson LW, Malinowski NM, Hyman R, Strong JM, Cysyk RL (November 1992). "Contribution of de-novo and salvage synthesis to the uracil nucleotide pool in mouse tissues and tumors in vivo". European Journal of Biochemistry. 210 (1): 293–6. doi:10.1111/j.1432-1033.1992.tb17420.x. PMID 1446677.